Running the pipeline
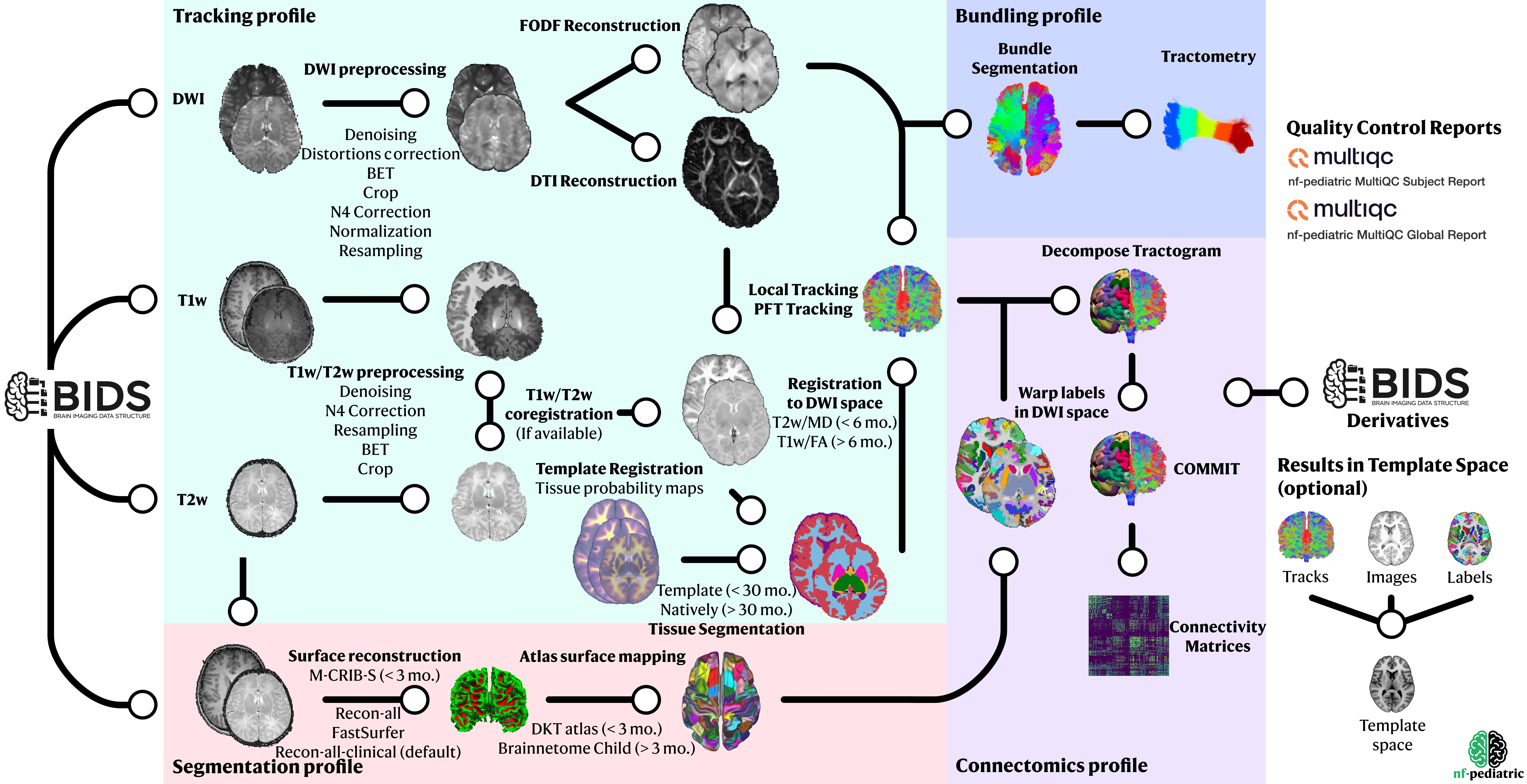
Choosing a profile
Section titled “Choosing a profile”nf-pediatric core functionalities are accessed and selected using profiles. This means users can select which part of the pipeline they want to run depending on their specific aims and the current state of their data (already preprocessed or not). As of now, here is a list of the available profiles and a short description of their processing steps:
Processing profiles:
-
-profile segmentation:By selecting this profile, FreeSurfer
recon-all, Recon-all-clinical, FastSurfer or M-CRIB-S/InfantFS will be used to process the T1w/T2w images and the Brainnetome Child Atlas (Li et al., 2022) or Desikan-Killiany (for infant (< 3 months)) will be registered using surface-based methods in the native subject space. -
-profile tracking:This is the core profile behind
nf-pediatric. By selecting it, DWI data will be preprocessed (denoised, corrected for distortion, normalized, resampled, …). In parallel, T1w will be preprocessed (if-profile segmentationis not selected), registered into diffusion space, and segmented to extract tissue masks/maps. Tissue segmentation method will be adapted based on the subject’s age. Preprocessed DWI data will be used to fit both the DTI and fODF models. As the final step, whole-brain tractography will be performed using both local tracking/particle filter tracking (PFT) and concatenated into a single tractogram. -
-profile bundling:This profile enables automatic bundle extraction from the processed whole-brain tractogram. By selecting it, bundle recognition will be performed in each subject using either the closest age-matched WM atlas (neonates, 3 months, 6 months, 12 months, 24 months, or children). Extracted bundles will then be filtered, uniformized, colored (affect only visualization), and tractometry will be performed to extract WM microstructure measures for each bundle.
-
-profile connectomics:By selecting this profile, labels will be registered in diffusion space and used to segment the tractogram into individual connections. The segmented tractogram will then be filtered, using COMMIT to remove false positive streamlines. Following filtering, connectivity matrices will be computed for a variety of metrics and outputted as numpy arrays usable for further statistical analysis.
Configuration profiles:
-
-profile docker(Recommended):Each process will be run using Docker containers.
-
-profile apptainer(Recommended):Each process will be run using Apptainer images.
-
-profile arm:Made to be use on computers with an ARM architecture (e.g., Mac M1,2,3,4). This is still experimental, depending on which profile you select, some containers might not be built for the ARM architecture. Feel free to open an issue if needed.
-
-profile slurm:If selected, the SLURM job scheduler will be used to dispatch jobs.
Using either -profile docker or -profile apptainer is highly recommended, as it controls the version of the software used and ensure reproducibility. While it is technically possible to run the pipeline without Docker or Apptainer, the amount of dependencies to install is simply not worth it.
Typical command-line
Section titled “Typical command-line”The typical command for running the pipeline is as follows:
nextflow run scilus/nf-pediatric -r 0.1.0 --input <BIDS_directory> --outdir ./results -profile docker,tracking -resumeThis will launch the pipeline with the docker configuration profile and will run the tracking processing steps. There are only 3 parameters that need to be supplied at runtime:
-
--input: for the path to your BIDS directoryFor more details on how to organize your input folder, please refer to the inputs section.
-
--outdir: path to the output directoryWe do not specify a default for the output directory location to ensure that users have total control on where the output files will be stored, as it can quickly grow into a large number of files. The recommended naming would be something along the line of
nf-pediatric-v{version}where{version}could be0.1.0for example. -
-profile: profile to be run and container system to usenf-pediatricprocessing steps was designed in profiles, giving users total control on which type of processing they want to make. One caveat is that users need to explicit tell which profile to run. This is done via the-profileparameter. To view the available processing profiles, please see this section -
-resume: Enablesnextflowcaching capabilities.This is a core
nextflowarguments. It enables the resumability of your pipeline. In the event where the pipeline fails for a variety of reasons, the following run will start back where it left off. For more details, see the corenextflowarguments section.
Note that the pipeline will create the following files in your working directory:
Directorywork/ # Directory containing the nextflow working files
- …
Directorynf-pediatric-v0.1.0/ # Finished results in specified location (defined with —outdir)
Directorymultiqc/
- …
- dataset_description.json
- README
Directorysub-01
Directoryses-01
Directoryanat/
- …
Directorydwi/
- …
Directoryfigures/
- …
Directorymultiqc/
- …
- .nextflow_log # Log file from Nextflow
- … # Other nextflow hidden files, eg. history of pipeline runs and old logs.
Using the params.yml file
Section titled “Using the params.yml file”If you wish to repeatedly use the same parameters for multiple runs, rather than specifying each flag in the command, you can specify these in a params file.
Pipeline settings can be provided in a yaml or json file via -params-file <file>.
The above pipeline run specified with a params file in yaml format:
nextflow run scilus/nf-pediatric -r 0.1.0 -profile docker -params-file params.yamlwith:
input: '<BIDS_directory>/'outdir: './results/'<...>To constrict the pipeline execution to specific subjects, you add the participant_label to the list:
input: '<BIDS_directory>/'outdir: './results/'participant_label: - sub-01 - sub-02<...>Reproducibility
Section titled “Reproducibility”It is a good idea to specify the pipeline version when running the pipeline on your data. This ensures that a specific version of the pipeline code and software are used when you run your pipeline. If you keep using the same tag, you’ll be running the same version of the pipeline, even if there have been changes to the code since.
First, go to the scilus/nf-pediatric releases page and find the latest pipeline version - numeric only (eg. 0.1.0). Then specify this when running the pipeline with -r (one hyphen) - eg. -r 0.1.0. Of course, you can switch to another version by changing the number after the -r flag.
This version number will be logged in reports when you run the pipeline, so that you’ll know what you used when you look back in the future. For example, at the bottom of the MultiQC reports.
To further assist in reproducibility, you can use share and reuse parameter files to repeat pipeline runs with the same settings without having to write out a command with every single parameter.
Core nextflow arguments
Section titled “Core nextflow arguments”-profile
Section titled “-profile”Use this parameter to choose a configuration profile. Profiles can give configuration presets for different compute environments.
Several generic profiles are bundled with the pipeline which instruct the pipeline to use software packaged using different methods (Docker, Singularity, and Apptainer) - see below.
The pipeline also dynamically loads configurations from https://github.com/nf-core/configs when it runs, making multiple config profiles for various institutional clusters available at run time. For more information and to check if your system is supported, please see the nf-core/configs documentation.
Note that multiple profiles can be loaded, for example: -profile tracking,docker - the order of arguments is important!
They are loaded in sequence, so later profiles can overwrite earlier profiles. For a complete description of the available profiles, please see this section.
-resume
Section titled “-resume”Specify this when restarting a pipeline. Nextflow will use cached results from any pipeline steps where the inputs are the same, continuing from where it got to previously. For input to be considered the same, not only the names must be identical but the files’ contents as well. For more info about this parameter, see this blog post.
You can also supply a run name to resume a specific run: -resume [run-name]. Use the nextflow log command to show previous run names.
Specify the path to a specific config file (this is a core Nextflow command). See the nf-core website documentation for more information.