Running the pipeline
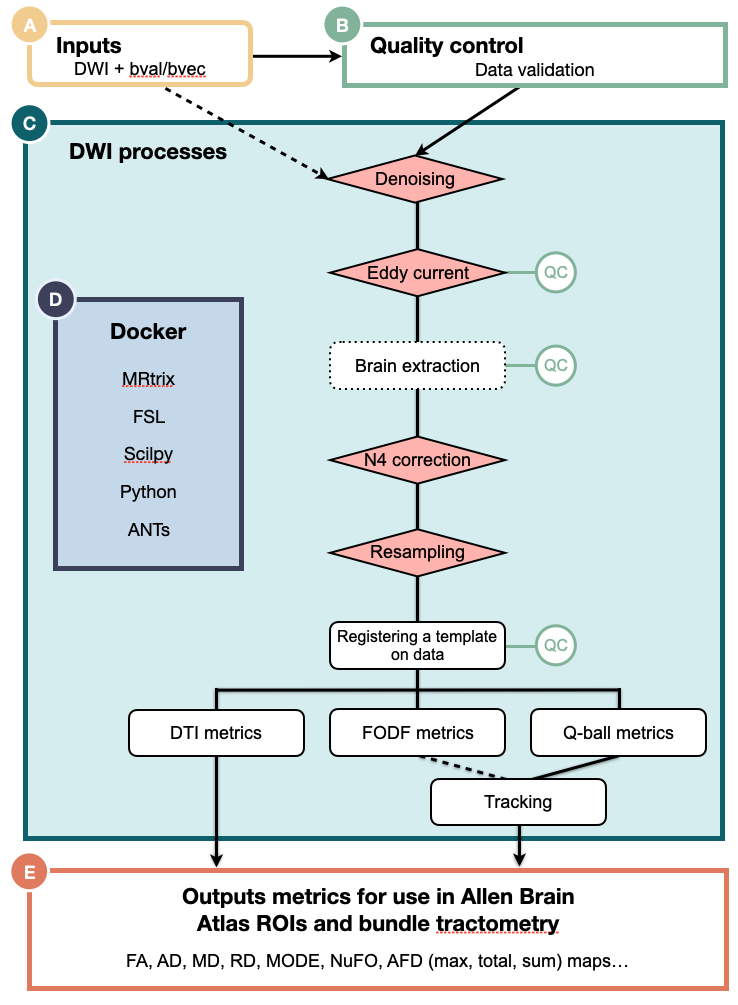
That said, here are some important parameters you might find useful.
Parameters
Section titled “Parameters”-
--use_preqcBy selecting this parameter and combined with the profile
run_preqc, you will use the output of a combination of checks and validations steps to insure your images and bvecs are oriented correctly. -
--use_fodf_for_trackingBy selecting this parameter, tracking will be carried out using the FODF reconstruction.
-
--run_stepEach preprocessing step of
nf-mousecan be skipped. You can deactivate a specific step by adding--run_STEP false. Here are the following steps that can be disabled: denoising, eddy, n4, resampling,
For the full parameter list, consult the page parameters
Profiles
Section titled “Profiles”Processing profiles:
-
pre_qc:By selecting this profile, you will run a set of checks and validations steps to insure your images and bvecs are oriented correctly.
Configuration profiles:
-
docker(Recommended):Each process will be run using Docker containers.
-
apptainer(Recommended):Each process will be run using Apptainer images.
-
arm:Made to be use on computers with an ARM architecture (e.g., Mac M1,2,3,4). This is still experimental, depending on which profile you select, some containers might not be built for the ARM architecture. Feel free to open an issue if needed.
-
slurm:If selected, the SLURM job scheduler will be used to dispatch jobs.
Using either -profile docker or -profile apptainer is highly recommended, as it controls the version of the software used and ensure reproducibility. While it is technically possible to run the pipeline without Docker or Apptainer, the amount of dependencies to install is simply not worth it.
Typical command-line
Section titled “Typical command-line”The typical command for running the pipeline is as follows:
nextflow run scilus/nf-mouse -r main \ --input <input_directory> \ --outdir <output_directory> \ -profile docker \ # or apptainer -with-report <report_name>.html \ -resumeThis will launch the pipeline using the main branch (latest non-released version) with the docker configuration profile.
There are only 3 parameters that need to be supplied at runtime:
-
--input: for the path to your BIDS directoryFor more details on how to organize your input folder, please refer to the inputs section.
-
--outdir: path to the output directoryWe do not specify a default for the output directory location to ensure that users have total control on where the output files will be stored, as it can quickly grow into a large number of files. The recommended naming would be something along the line of
nf-mouse-v{version}where{version}could be0.1.0for example. -
-r: Release nf-mouseYou need to select the
nf-mouserelease you want to use. Release available. -
-profile: profile to be run and container system to usenf-mouseprocessing steps was designed in profiles, giving users total control on which type of processing they want to make. One caveat is that users need to explicit tell which profile to run. This is done via the-profileparameter. To view the available processing profiles, please see this section -
-resume: Enablesnextflowcaching capabilities.This is a core
nextflowarguments. It enables the resumability of your pipeline. In the event where the pipeline fails for a variety of reasons, the following run will start back where it left off. For more details, see the corenextflowarguments section. -
-with-report: Enablesnextflowcaching capabilities.This is a core
nextflowarguments. It enables the html report of your pipeline. For more details, see the corenextflowarguments section.
Now it’s time to look at your output directory. See output section.
Build your own command line
Section titled “Build your own command line”Here is a smart tool to help you build
Advanced usage
Section titled “Advanced usage”The execution method for nf-mouse varies, often depending on the informatics structure where you intend to run nf-mouse.
Here are three extra sections that could help you:
Outputs
Section titled “Outputs”You are now ready to explore the output BIDS structure.

